Protein Biology Takes a Giant Leap Into the Future
The genome holds instructions for creating and maintaining an organism, but most physiological functions involve what genes are translated into - proteins. Every cell holds the proteins that give it an identity and enable it to do its job, and all of those thousands of proteins have to work together in carefully coordinated interactions. When problems arise in proteins, it leads to disease, so it's essential to understand the structure and function of proteins in order to understand and treat disease.
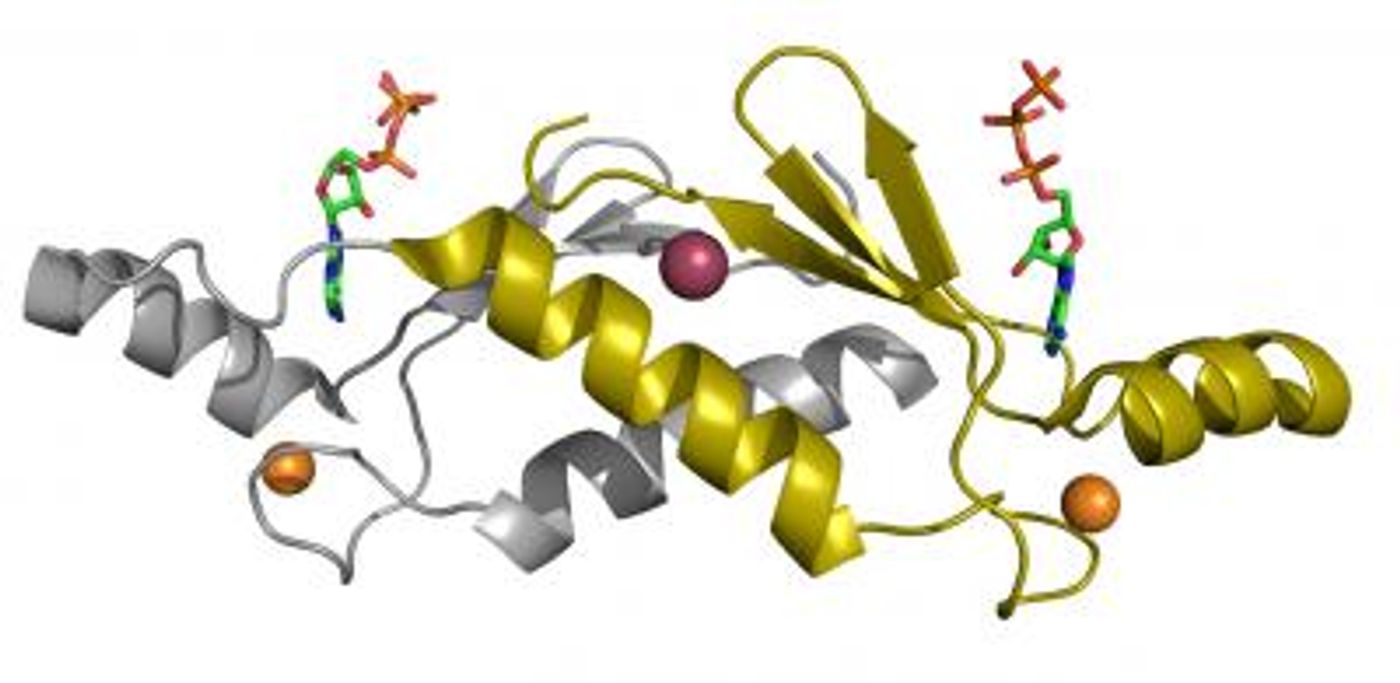
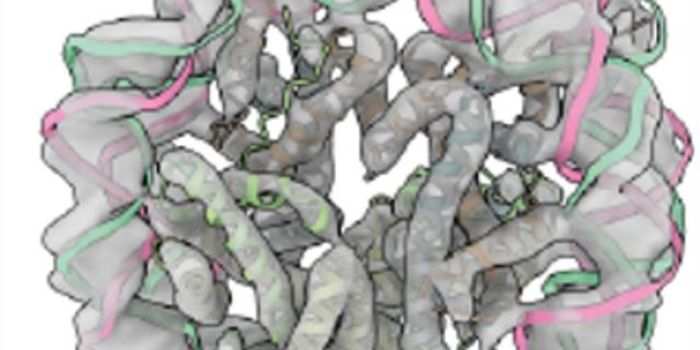
A gene sequence is transcribed into RNA, then modified before being translated by cellular machinery into a string of amino acids. These amino acid strings are then processed in cellular organelles and carefully folded into a three-dimensional structure. Therefore, a protein's structure is not immediately evident from its sequence, and it used to take time-consuming, painstaking work to decipher it.
New work has propelled protein research forward with the creation of a tool that can accurately predict the structure of a protein using only its sequence. Hundreds of researchers worked together to create an artificial intelligence (AI) program called AlphaFold. The work was announced at the 14th Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction (CASP14).
"Proteins are extremely complicated molecules, and their precise three-dimensional structure is key to the many roles they perform, for example the insulin that regulates sugar levels in our blood and the antibodies that help us fight infections. Even tiny rearrangements of these vital molecules can have catastrophic effects on our health, so one of the most efficient ways to understand disease and find new treatments is to study the proteins involved," said the chair of CASP14, Dr. John Moult of the University of Maryland.
"There are tens of thousands of human proteins and many billions in other species, including bacteria and viruses, but working out the shape of just one requires expensive equipment and can take years," added Moult.
In this work, the researchers built on CASP13, created 2018. The AI lab DeepMind trained a neural network system that interprets a spatial graph representing a protein. These spatial graphs help define the interactions between different parts of a single protein. The program was trained using publicly available data on about 170,000 protein structures, which has allowed AlphaFold to make predictions about new protein structures.
"What the DeepMind team has managed to achieve is fantastic and will change the future of structural biology and protein research. After decades of studying proteins, the molecules that provide the structure and functions of all living things, I awoke this morning feeling that progress has been made," said Professor Dame Janet Thornton, Director Emeritus & Senior Scientist at EMBL-EBI.
"AlphaFold's astonishingly accurate models have allowed us to solve a protein structure we were stuck on for close to a decade, relaunching our effort to understand how signals are transmitted across cell membranes," said Professor Andrei Lupas, Director of the Max Planck Institute for Developmental Biology.
"I nearly fell off my chair when I saw these results. I know how rigorous CASP is - it basically ensures that computational modelling must perform on the challenging task of ab-initio protein folding. It was humbling to see that these models could do that so accurately. There will be many aspects to understand but this is a huge advance for science," commented Professor Ewan Birney, Deputy Director-General of EMBL, and Director of EMBL-EBI.
Sources: AAAS/Eurekalert! via Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction, Nature, DeepMind