Methylation Affects the 3D Structure of the Genome
Gene activity has to be carefully controlled by cells so that they maintain their identity and continue to carry out their proper functions, and there are many different ways that gene expression can be influenced and altered. Epigenetic features of the genome are just one aspect of gene regulation and they exert their influence without making any changes to the sequence of the DNA.
Another aspect of gene regulation is the three-dimensional structure of the genome. When genes are active, the cellular machinery that transcribes genes has to be able to physically contact that portion of the genome so it can be copied into an RNA molecule. Methyl groups, chemical tags that can be added to the genome, are yet another aspect of gene expression. It's been suggested, for example, that the presence of many methyl groups in certain places might suppress gene expression.
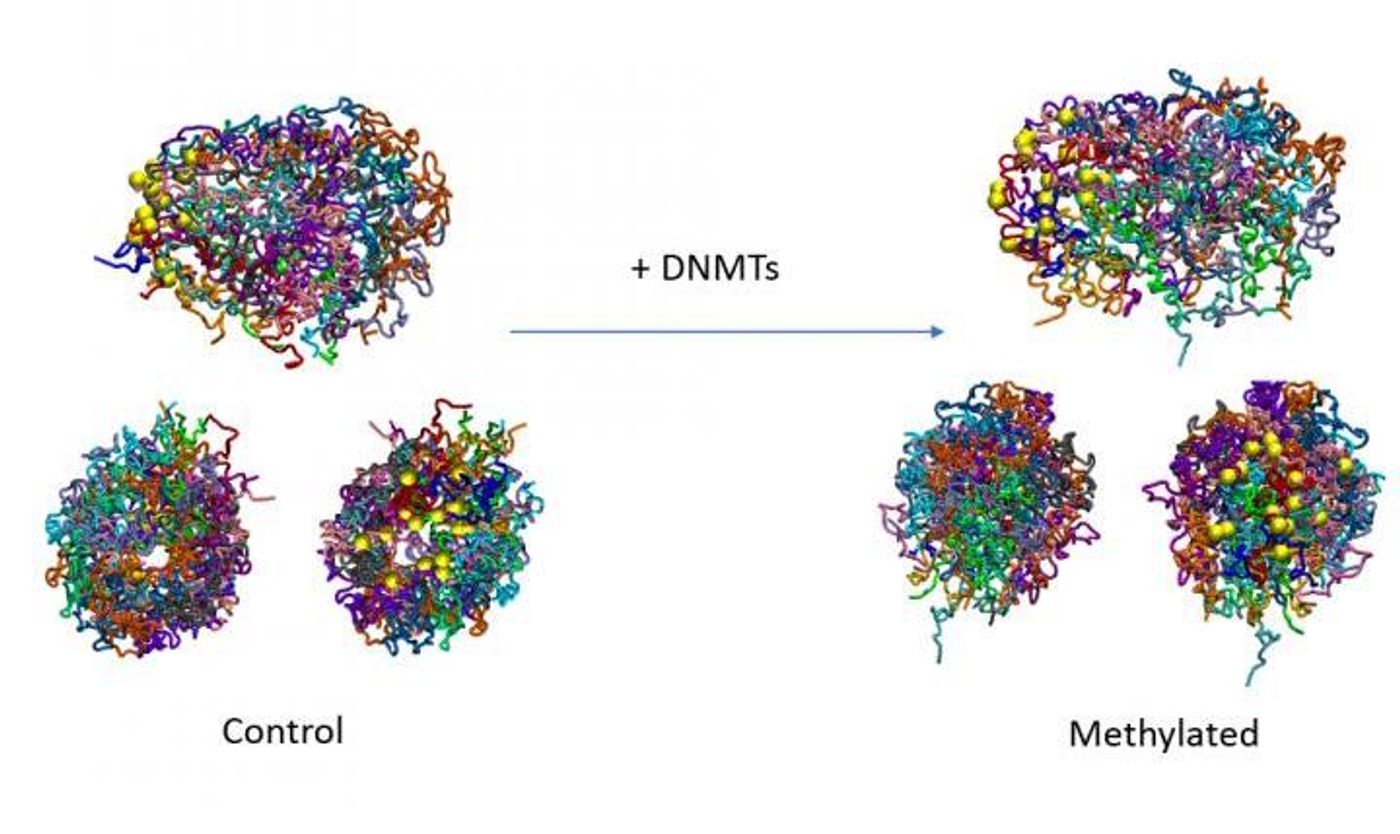
A new study has also shown that the relationship between methyl groups and gene activity may be even more complex than we thought; it seems that methylation can change the three-dimensional structure of the genome.
Reporting in Nature Communications, researchers used yeast cells to show that methyl groups can increase the stiffness of the DNA molecule, which influences the structure and thus, alters gene expression. This mechanism illustrates how closely epigenetic features are linked to gene regulation.
In this study, the researchers utilized budding yeast, which doesn't have the cellular machinery to add methyl groups to its genome. The scientists expressed mouse versions of enzymes called DNA methyltransferases to see how the yeast genome, which did not naturally carry methyl groups, reacted when methyl groups were added to it. The investigators showed that genes that were active displayed varying levels of methylation at the places where transcription started and ended, like what's seen in mammalian cells.
"The new model organism and the theoretical analysis framework that we have developed and published are really innovative and we hope they will facilitate research projects undertaken by many laboratories around the world studying DNA methylation and its impact on gene expression," noted study leader Dr. Modesto Orozco, head of the Molecular Modelling and Bioinformatics Lab at IRB Barcelona.
Computer modeling and simulations were essential to this study. "Using these techniques, we observed that we could recapitulate the characteristic distribution of DNA methylation seen in mammalian genomes, and we confirmed our earlier in vitro result on the relationship between 3D structure, DNA flexibility, and methylation, showing that this also occurs in vivo," explained study co-director Dr. Isabelle Brun Heath, director of the Experimental Bioinformatics Laboratory.
Sources: AAAS/Eurekalert! via Institute for Research in Biomedicine (IRB Barcelona), Nature Communications