Within Cells, DNA can Move Molecules
While DNA is portrayed as a beautiful double helix, in live cells it’s a clump of tangles that seems impenetrable. However, there are lots of important molecules that need to get to the right parts of specific areas of DNA, and scientists wondered how they could do that in such a dense cluster. New work published in Biophysical Journal by researchers at the Georgia Institute of Technology has indicated that DNA is not immobile but has dynamic properties that allow it to push those critical molecules to the right places at the right times.
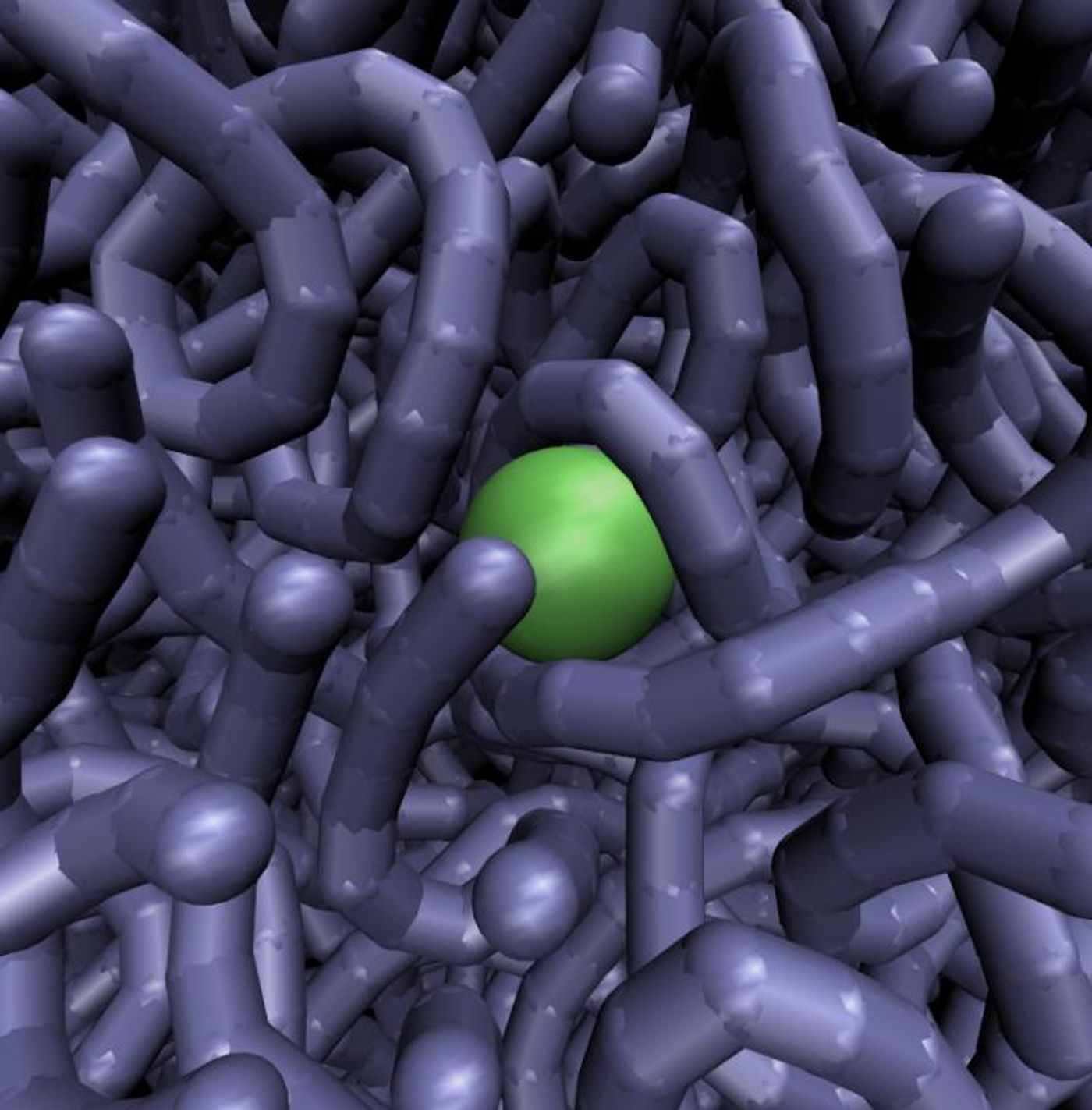
Transcription factors work to express the proper genes, and they are very large molecules. It’s long been a mystery as to how they got where they needed to be in the dense thicket that is natural DNA. In the video above, the green ball represents a transcription factor within grey DNA strands.
"If the thicket is so thick, and on top of that doesn't move, then it should be impenetrable. So, how do you get stuff through to the right site?" queried Jeffrey Skolnick, a Professor at Georgia Tech's School of Biological Sciences.
Skolnick collaborated with a computer scientist that specializes in algorithms for scientific inquiries, Edmond Chow. They challenged the idea that DNA is a rigid structure, and instead simulated DNA as a moveable molecule that can twist and turn moving itself and the stuff around it.
"The DNA motion is far and away the dominant force moving molecules through its thicket," Skolnick explained. "DNA is a bully."
For this work the investigators used the well-known transcription factor LacI (moedled in the videos as the green ball) from the bacterium Esherichia coli for their study. In the computer simulations, strands of DNA could move out of the way of LacI and also propel the LacI forward to the next space. This could help explain why transcription factors seem to move so slow in simulations that use DNA that does not flex. When the DNA wiggles, the LacI can speed along.
Transcription factors are known to be able to hop on and off, clicking into the right spot on DNA when needed. "But the sliding and hopping combined still don't account for the speed of diffusion," Chow noted. However, the DNA can flick the transcription factor as it hops, propelling it ahead and increasing their diffusion rate.
The revelations in this work were made possible by massive computational power. "These simulations are unique to this problem because of their enormity and the advanced computing techniques used. Very efficient algorithms ran in parallel on powerful computers, and, still, it took three weeks for the simulations to complete," Chow commented.
It still required some trimming of real-life conditions in order to be able to run the entire simulation. Scientists are still working to develop models that can account for the entire framework of the cell. "The ultimate goal is to put a whole cell on a computer. Let it live. Let it divide, and understand the processes," Skolnick said. "Maybe even let the cell mutate and evolve."
"When the size of a problem grows, the computing costs to solve it can grow disproportionately," Chow noted. "You have to build algorithms that can run efficiently even when you scale up the problem size."
Sources: AAAS/Eurekalert! via Georgia Institute of Technology, Biophysical Journal