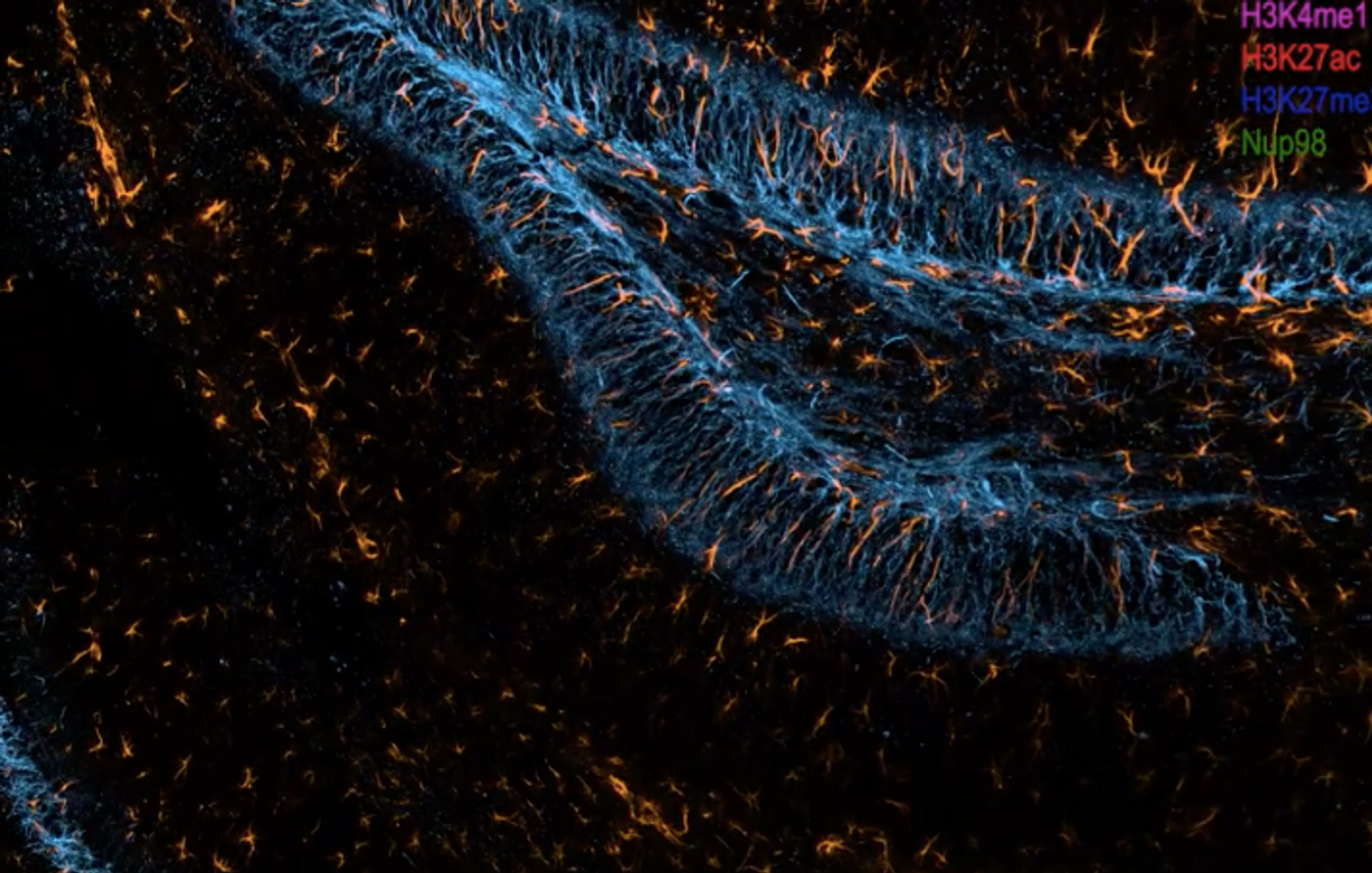
Cutting-Edge Tech Can ID RNA & Protein in Deep Tissues
Genes have to be expressed in the right place, at the time, and at the right levels to carry out the right functions. To learn more about these physiological processes and how they change in disease, scientists want to track that gene and protein expression. But to do this with thick tissue samples, a new technique had to be developed that would enable researchers to monitor the expression of genes as messenger RNA (mRNA) and then as the proteins that mRNA encodes for. This method, called cycleHCR, barcodes and colors RNA sequences and proteins so they can be seen with fluorescence microscopy. The work has been reported in Science.
CycleHCR uses a previously created tool called Hybridization Chain Reaction, or HCR. Multiple colors or fluorophores can be added to a target sequence, so they can be visualized even when they sit deep inside of tissue blocks. Since there are only so many fluorophores, this technique was limited, and only about four colors could be seen at any one time. But there are many more molecules in tissues.
The scientists created new DNA barcodes that could be linked to targets, and in this case, every molecule could be tagged in a unique way. The barcodes have two parts, so they are specific enough to identify certain mRNA molecules. The target can also be amplified through HCR when found.
These barcodes are also easy to remove, so HCR can be done on one sample many times. One round of imaging focuses on three barcodes, and three kinds of RNA molecules. These barcodes are then removed. The next round does a different set of three. This process can be repeated many times to detect a virtually unlimited number of targets in one sample.
“I think it will be a gamechanger very broadly, not just for people in my field,” said senior study author James Liu, a group leader at Howard Hughes Medical Institute's Janelia Research Campus. “It was a tool developed to answer a very obscure question, but I think all biologists can use the technique in their favorite samples.”
“We modified the split amplification chain reaction technique in a way that now we are adding barcoding to it where we can detect hundreds, potentially even thousands of RNAs, with these multi rounds,” added co-first study author Valentina Gandin, a Senior Scientist in the Liu Lab. “The barcoding was a novelty that we added to this.”
These barcodes can detect RNA, but this same approach was also then used to detect proteins; so the spatial and temporal organization of both RNA transcripts and proteins can be analyzed.
Since this method requires many repeat steps, the investigators also automated it, so gene expression can be mapped in tissues, and researchers can assess the data more easily.
The team used the tool to measure and observe the expression of 254 genes in one mouse embryo sample. All of the cell types in this sample were easily characterized, and new cell types were discovered.
Since the barcode sequencing and most aspects of the method are freely available, the researchers are hopeful that this technique will spur new discoveries. The team is also hoping to share the automation protocols as well.
“Eventually we want everybody to use it,” said Liu. “We would really like our technique to be broadly spread to enable every scientist to be able to use it.”
Sources: Howard Hughes Medical Institute, Science