Same Repetitive DNA Sequence at the Root of Several Rare Neurodegenerative Disorders
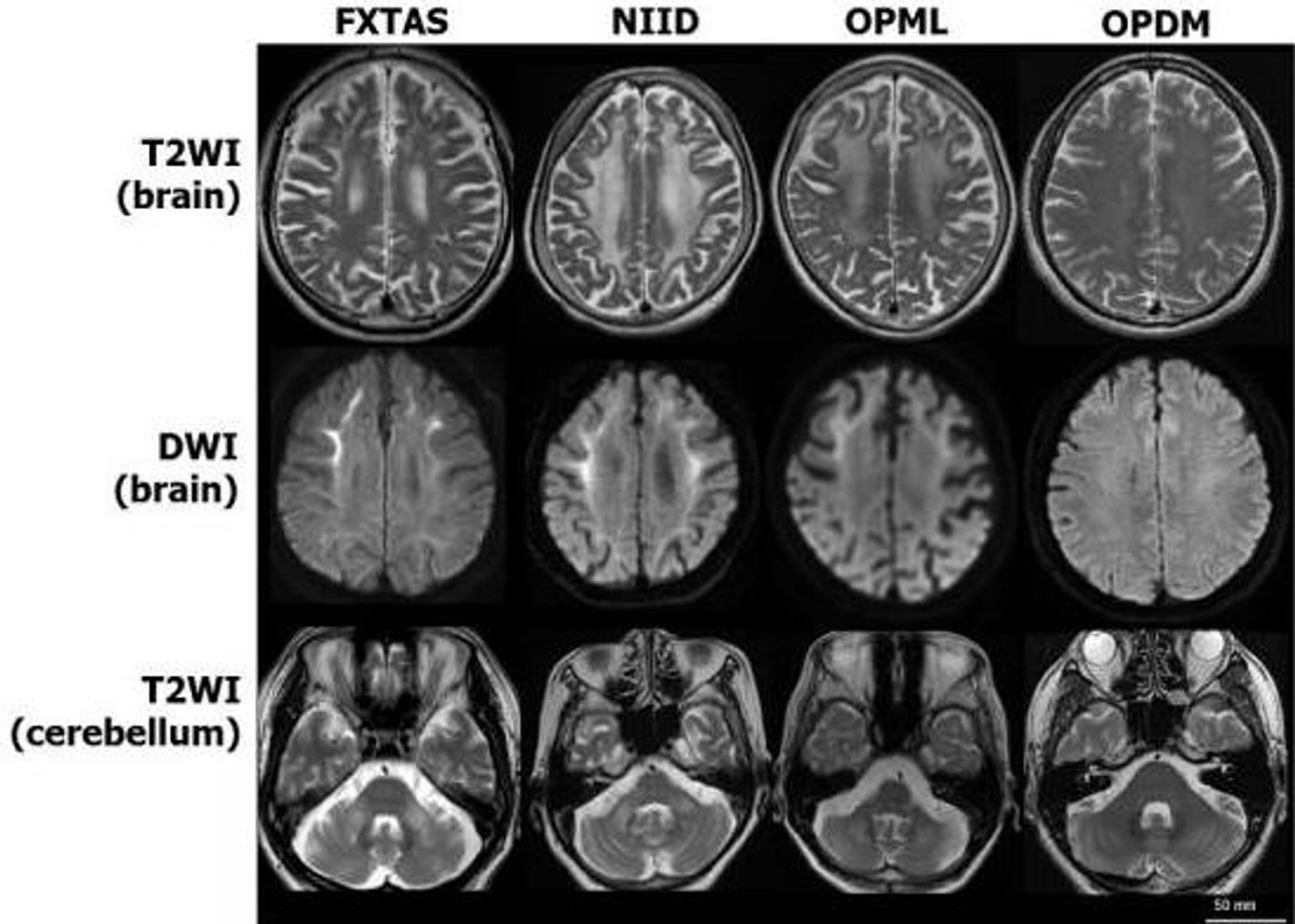
Reporting in Nature Genetics, scientists have now identified a genetic feature underlying three neurodegenerative diseases; these rare disorders are caused by a repetitive series of DNA bases. The illnesses, neuronal intranuclear inclusion disease, oculopharyngodistal myopathy, and an overlapping disease, have some symptoms in common with fragile X ataxia/tremor syndrome. That disease is caused by an excessive repetition of the DNA base sequence CGG too many times - one example of a tandem repeat mutation, in this case on the X chromosome. The researchers suspected that the same mutation may also underlie the other diseases.
"Because the mutations causing the diseases are so similar, in the future, all these patients might benefit from the same treatment," explained Dr. Hiroyuki Ishiura, M.D./Ph.D., an assistant professor from the University of Tokyo Hospital and study first author.
Now the researchers can pursue a therapeutic aimed at the tandem repeat, which could apply to the various disorders the tandem repeat mutation can cause, rather than a specific, rare disorder needing a tailored therapy. It is unclear, however, whether the researchers modeled any of the mutations they identified in an organism in an attempt to recapitulate the symptoms.
"Gene silencing techniques [that keep genes inactive] are a possible treatment. We cannot know the result, but we believe such strategies may help patients in the future," said the corresponding author of the report Shoji Tsuji, MD, Ph.D., project professor from the University of Tokyo Hospital.
The neurodegenerative diseases cause uncontrolled movements, cognitive impairment, weakness in the limbs, or swallowing difficulties. In the case of fragile X ataxia/tremors, the CGG repeats causing the illness are located on the X chromosome, but the rare disease patients had normal X chromosomes. A wide search for other repetitive sequences would even recently have been extremely laborious and very challenging (tandem repeats can be of any bases - A, T, G or C, repeated any unusual number of times on any chromosome). Current genome sequencing techniques are much faster, and computational analysis tools are more powerful, however.
The researchers sequenced the patient genomes and some healthy controls in short stretches, then sorted through the data to look for repeats of CGG, and find where in the genome they were located. They focused on the places where patients carried many CGG repeats that healthy people didn’t have and sequenced longer stretches of those regions. The different diseases probably trace back to CGG repeats in seemingly unrelated portions of the genome.
The researchers have now provided one family with an answer to the question of the cause of the disease they carry. The team is analyzing more genomes to find rare cases of healthy individuals that carry CGG repeats. They also want to learn more about whether tandem repeats play a role in other neurodegenerative disorders with uncertain causes.
Learn more about how repetitive sequences may play a role in disease from the video above by the Broad Institute.
Sources: AAAS/Eurekalert! via University of Tokyo, Nature Genetics