Pinpointing the Location of Gene Expression in Various Tissues
Researchers can easily assess what genes are being expressed in a tissue by using a method like RT-PCR that precisely quantifies gene activity, or western blots that identify protein levels. But those tools use tissue that has been ground up and chemically treated so that RNA or proteins can be isolated; a different technique has to be used to look for where genes are being expressed in solid tissues. Scientists have created tools that can now illuminate gene expression throughout tissues, like organs or tumors, with precision. Patterns of gene activity can now be visualized along with the identity of crucial proteins in various tissue samples. The technique has been reported in Nature Biotechnology, and can be used to map gene expression in tissues and tumors, crucial knowledge for basic research.
“This technology is exciting because it allows us to map the spatial organization of tissues, including cell types, cell activities and cell-to-cell interactions, as never before,” said co-senior study author Dr. Dan Landau, an associate professor of medicine at Weill Cornell Medicine, among other appointments.
This method is called Spatial PrOtein and Transcriptome Sequencing (SPOTS). It utilizes glass slides that are for imaging, but have also been coated with special probes that contain molecular barcodes that indicate a spatial location. Thin slices of tissue are placed on the slide, and the cells are treated so that the probes can attach to messenger RNA (mRNA) molecules in the cells, which are indicative of genes that are being expressed. Antibodies are used to bind to important proteins and the spatial probes. Captured mRNAs and proteins are automatically identified and mapped to their locations within the sample using computational tools. The resulting data might be directly analyzed, or compared to pathological samples.
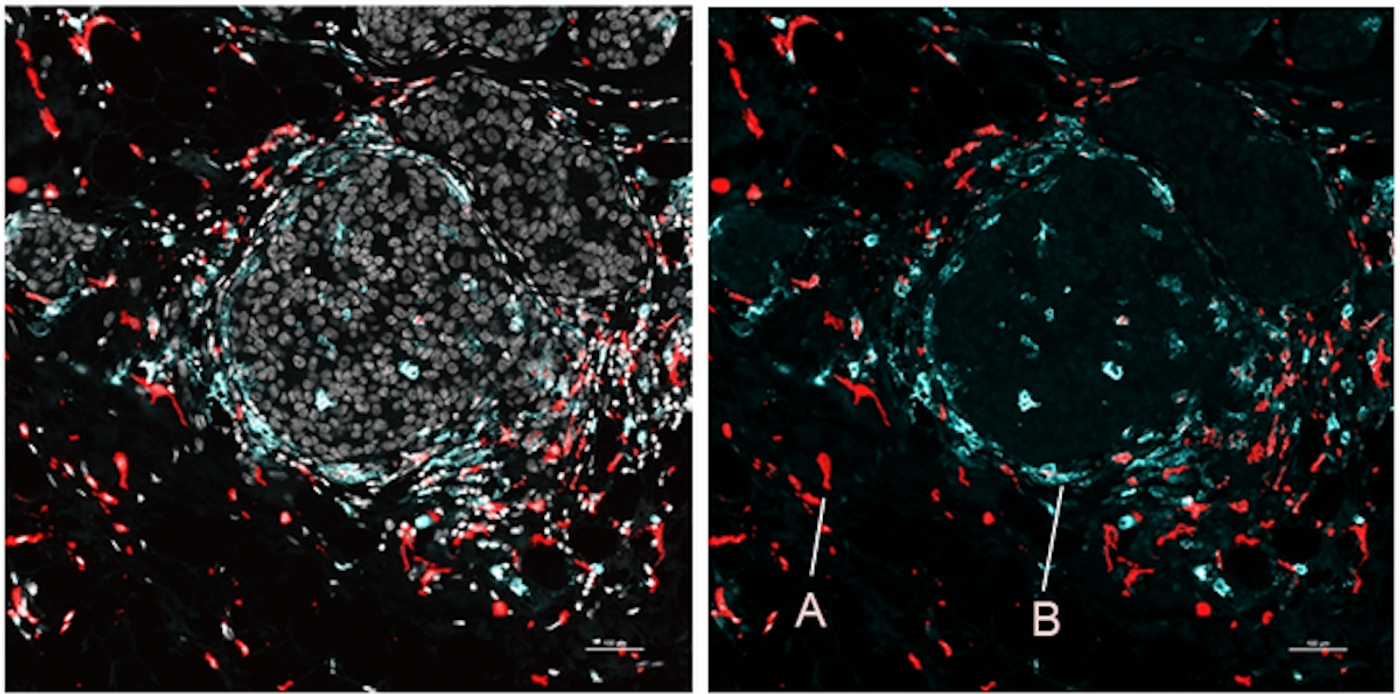
The researchers showed that SPOTS worked on normal tissue from a mouse spleen, and revealed the organ's functional construction, such as where different types of cells were clustered and working throughout the organ.
SPOTS was also used to assess breast tumor tissue from a mouse model. Immune cells were shown in the tissue in two states, one of which formed a protective, immunosuppressive barrier while another battled the tumor. Those two types of cells were found in different parts of the tumor, interacting with other, different cells; those two states are probably driven by the tumor microenvironment, noted Landau.
This type of tool could show why some patients can be helped with immunotherapeutics whole others don't respond to it. It may also lead to the creation of improved therapeutics, added Landau.
While this method cannot yet resolve gene expression in tissues at the single-cell level, and instead shows information for a few cells together, the researchers are hopeful that they will soon drill down to the level of individual cells with SPOTS, and include other data as well.
Sources: Weill Cornell Medicine, Nature Biotechnology