A simplified high-throughput protein expression system
Protein synthesis, protein engineering, and synthetic biology researchers can research their protein of interest using either in vivo or in vitro protein expression systems. Each system has its upsides, but in vitro expression systems are best suited for high-throughput protein expression applications.
In vivo protein expression
In vivo protein expression includes cloning a gene of interest with the essential control elements into an expression vector, and then transforming the vector into an easily grown organism or cell such as bacteria, yeast, or cultured cells. After transcription and translation, the protein is purified from the rest of the cellular materials.
The advantages of in vivo expression systems include using native enzymes and modification systems. Plus, once the recombinant cell is established, there is an endless supply of the protein. However, this is a low-throughput strategy that can take many days to complete and is often fraught with physiological constraints such as in vivo toxicity, protein insolubility, substrate competition, and the inability to incorporate nonnatural amino acids [1,2].
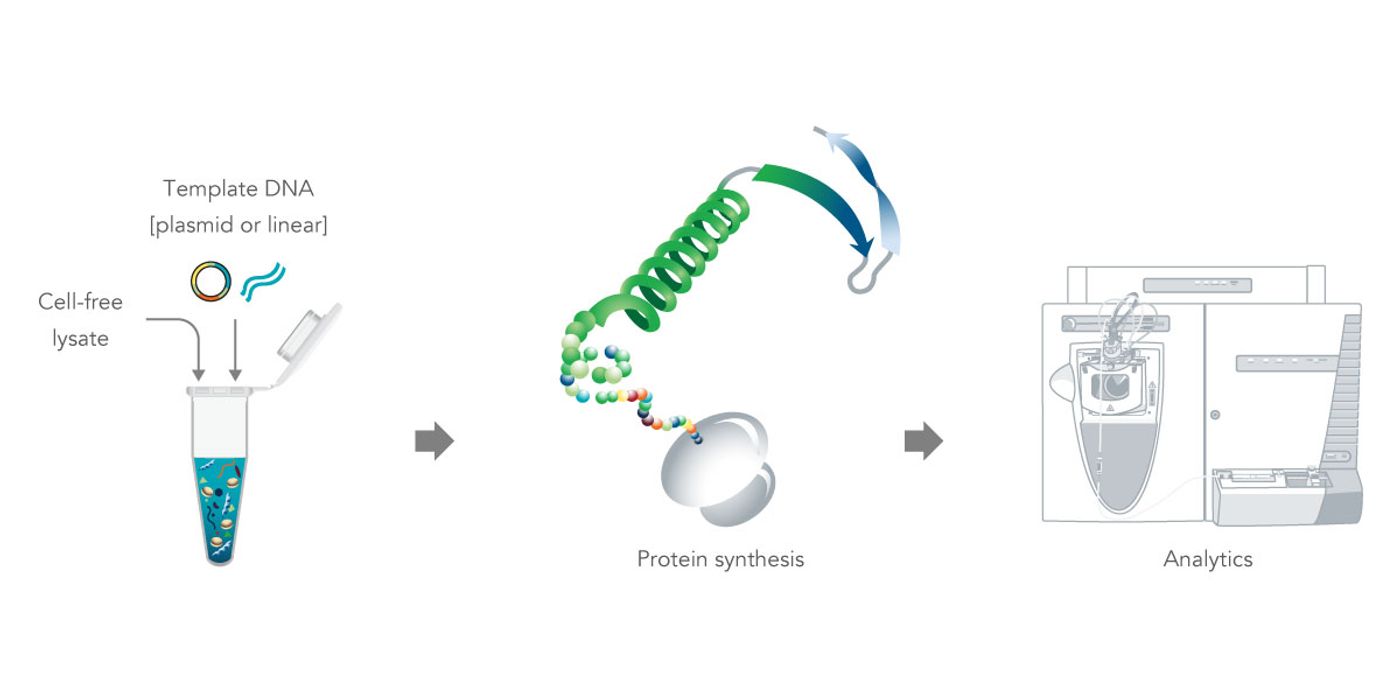
Figure 1. Cell-free protein expression systems. Combining cell-free lysates and template DNA provides a convenient method for in vitro protein synthesis for high-throughput applications.
What is cell-free expression?
Cell-free protein synthesis circumvents and simplifies protein production workflows. Although the technique is not new, recent improvements have made it more convenient, especially in high-throughput applications.
In vitro protein expression requires two components (Figure 1). First, cellular extracts are required. These contain a specific blend of essential enzymes for RNA transcription and protein translation, amino acids, ribonucleotides, tRNA, co-factors, ATP, and buffers. These cellular lysates are available in user-friendly, master mix formulations [3]. Cellular lysates for in vitro protein transcription and translation are derived from prokaryotes such as E. coli, insect cells, or eukaryotic cells such as wheat germ or rabbit reticulocytes [2], which offer flexibility for modification types.
The second component is a DNA template with a 5’ untranslated region (UTR), promoter, other gene expression control elements, protein coding sequence, transcriptional terminator, and 3’ UTR. The simplest method of creating the template DNA is to order a custom linear double-stranded DNA (dsDNA) gene fragment or gene, both available through Integrated DNA Technologies. The entire template can be synthesized as a single dsDNA fragment since the fragments are available in lengths up to 3000 bp. There is no need to manipulate the DNA with PCR, subcloning fragments, vector construction, or DNA extractions. Simply rehydrate your custom designed fragment, add to the master mix, incubate, and test the expressed protein without purification.
Benefits of in vitro transcription and translation
Besides speed, cell-free transcription and translation provides many benefits [4]:
- Flexible template design. A recent application note, “An accelerated protein engineering workflow utilizing linear DNA in a cell-free expression system”, found that there is a large amount of flexibility for the lengths of 5’- and 3’-flanking regions for dsDNA templates.
- High-throughput proteins. In vitro protein production works with liquid handling systems (multiwell plate format or large aliquot sizes are available).
- Protein folding modifications. Adding purified scaffolding proteins or folding proteins can improve the final structure of the protein of interest.
- Expanded genetic code with unnatural amino acids. Cell-free master mixes can contain unnatural amino acids for protein structure analysis methods.
- Post-translational modifications. Disulfide bond formation, glycosylation, phosphorylation, ubiquitination, and other modifications are possible with augmented cell lysates.
Whether creating protein libraries for functional screens, performing protein evolution, antibody fragment binding studies, or producing proteins to research potential therapeutics, cell-free protein expression provides a simple high-throughput approach that saves time and improves your discovery pipeline.
To learn more about how to streamline your research timelines, watch the IDT on-demand webinar, “Accelerate your protein expression workflows.”
- Khambhati K, Bhattacharjee G, Gohil N, et al. Exploring the Potential of Cell-Free Protein Synthesis for Extending the Abilities of Biological Systems. Front Bioeng Biotechnol. 2019;7:248.
- Gregorio NE, Levine MZ, Oza JP. A User's Guide to Cell-Free Protein Synthesis. Methods Protoc. 2019;2(1).
- Garamella J, Marshall R, Rustad M, et al. The All E. coli TX-TL Toolbox 2.0: A Platform for Cell-Free Synthetic Biology. ACS Synth Biol. 2016;5(4):344-355.
- Silverman AD, Karim AS, Jewett MC. Cell-free gene expression: an expanded repertoire of applications. Nat Rev Genet. 2020;21(3):151-170.